Overview
We have several ongoing research projects in the areas outlined below. Approaches range from genomics (dry-lab) to functional experimentation (wet-lab) to clinical/translational efforts. Please reach out if you are interested in joining or collaborating with the lab.
Research Motivation
The genomics revolution has given us the power to understand the molecular wiring of cancer cells in unprecedented detail and has enabled the development of highly effective targeted therapies that perturb specific molecular pathways essential to the cancer cell. Still, there are many gaps in our understanding of how cancer develops and progresses. For example:
We are interested in exploring these and other fundamental questions in cancer biology using multidisciplinary and collaborative approaches, with the ultimate goal of laying the pre-clinical foundation for the next generation of targeted therapies that can improve outcomes for cancer patients.
We have several ongoing research projects in the areas outlined below. Approaches range from genomics (dry-lab) to functional experimentation (wet-lab) to clinical/translational efforts. Please reach out if you are interested in joining or collaborating with the lab.
Research Motivation
The genomics revolution has given us the power to understand the molecular wiring of cancer cells in unprecedented detail and has enabled the development of highly effective targeted therapies that perturb specific molecular pathways essential to the cancer cell. Still, there are many gaps in our understanding of how cancer develops and progresses. For example:
- What are the significant drivers of cancer that occur in non-coding or “dark” regions of the genome, and how can these be rationally targeted?
- What are the drivers of rare cancers that have not been comprehensively profiled?
- How do cancer genomes and transcriptomes evolve during treatment?
We are interested in exploring these and other fundamental questions in cancer biology using multidisciplinary and collaborative approaches, with the ultimate goal of laying the pre-clinical foundation for the next generation of targeted therapies that can improve outcomes for cancer patients.
Research Areas
Genetics and biology of genitourinary cancers
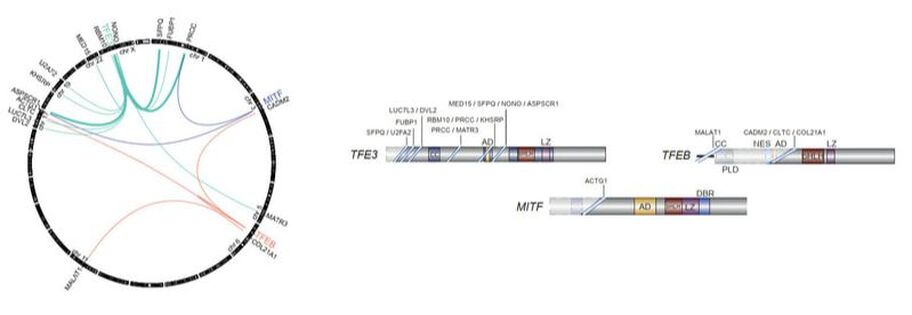
 Diverse alterations involving the androgen receptor locus in castration-resistant prostate cancer (from Viswanathan, Ha, Hoff et al. Cell, 2018)
Diverse alterations involving the androgen receptor locus in castration-resistant prostate cancer (from Viswanathan, Ha, Hoff et al. Cell, 2018)
1. Charting the Molecular Landscape of Genitourinary Cancers
Recent large-scale sequencing efforts have been instrumental in delineating the most significantly mutated cancer genes across many common cancer types. Still, most of these efforts have primarily interrogated coding sequences, which constitute only a small fraction (<2%) of the genome. Many alterations in noncoding or poorly mappable regions of the cancer genome may hence remain to be discovered.
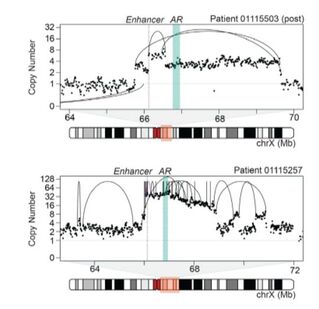
Prostate Cancer: Using linked-read whole genome sequencing, we recently described highly-recurrent duplications involving a novel long-range enhancer of the androgen receptor (AR) in > 80% of cases of castration-resistant prostate cancer. These pervasive alterations play an important role in mediating resistance to anti-androgen therapies.
MiT/TFE translocation renal cell carcinoma (tRCC): Our laboratory has a special interest in the genetics and biology of tRCC, a rare but highly aggressive subtype of kidney cancer without effective therapies. This cancer is driven by oncogenic gene fusions involving a transcription factor in the MiT/TFE family, usually TFE3 located on chromosome Xp11.2. Through a large-scale genomic effort, we have recently described the molecular and clinical landmarks of tRCC.
Recent large-scale sequencing efforts have been instrumental in delineating the most significantly mutated cancer genes across many common cancer types. Still, most of these efforts have primarily interrogated coding sequences, which constitute only a small fraction (<2%) of the genome. Many alterations in noncoding or poorly mappable regions of the cancer genome may hence remain to be discovered.
Prostate Cancer: Using linked-read whole genome sequencing, we recently described highly-recurrent duplications involving a novel long-range enhancer of the androgen receptor (AR) in > 80% of cases of castration-resistant prostate cancer. These pervasive alterations play an important role in mediating resistance to anti-androgen therapies.
MiT/TFE translocation renal cell carcinoma (tRCC): Our laboratory has a special interest in the genetics and biology of tRCC, a rare but highly aggressive subtype of kidney cancer without effective therapies. This cancer is driven by oncogenic gene fusions involving a transcription factor in the MiT/TFE family, usually TFE3 located on chromosome Xp11.2. Through a large-scale genomic effort, we have recently described the molecular and clinical landmarks of tRCC.
 Top: Paralog vulnerabilities identified from analysis of genome-scale genetic screening of cancer cell lines by shRNA (top-left) or CRISPR (top-right).
Bottom: Transcriptomic changes seen upon MAGOHB knockdown in either a MAGOH-deleted (bottom-left) or MAGOH-replete (bottom-right) genetic context. From Viswanathan et al., Nature Genetics, 2018
Top: Paralog vulnerabilities identified from analysis of genome-scale genetic screening of cancer cell lines by shRNA (top-left) or CRISPR (top-right).
Bottom: Transcriptomic changes seen upon MAGOHB knockdown in either a MAGOH-deleted (bottom-left) or MAGOH-replete (bottom-right) genetic context. From Viswanathan et al., Nature Genetics, 2018
2. Identifying Novel Molecular Vulnerabilities of Genitourinary Cancers
The capacity to integrate genomic and transcriptomic features of cancer cell line models with functional genetic perturbation performed at genome-scale has yielded an unprecedented ability to precisely map the molecular contexts in which cancers display specific (and often unexpected) molecular vulnerabilities.
Paralog lethality across cancers: Redundant essentiality between paralog genes may be therapeutically exploited when one of two copies of an essential paralog pair is somatically deleted in cancer. We have recently reported several such paralog pairs, including the example of MAGOHB, a component of the exon-junction complex that proves essential to maintaining RNA surveillance when its paralog, MAGOH, is somatically deleted. By performing genome-scale genetic screening in models of genitourinary cancer, we aim to uncover novel and therapeutically tractable molecular targets that can inspire the development of the next-generation of therapies for these cancers.
2. Identifying Novel Molecular Vulnerabilities of Genitourinary Cancers
The capacity to integrate genomic and transcriptomic features of cancer cell line models with functional genetic perturbation performed at genome-scale has yielded an unprecedented ability to precisely map the molecular contexts in which cancers display specific (and often unexpected) molecular vulnerabilities.
Paralog lethality across cancers: Redundant essentiality between paralog genes may be therapeutically exploited when one of two copies of an essential paralog pair is somatically deleted in cancer. We have recently reported several such paralog pairs, including the example of MAGOHB, a component of the exon-junction complex that proves essential to maintaining RNA surveillance when its paralog, MAGOH, is somatically deleted. By performing genome-scale genetic screening in models of genitourinary cancer, we aim to uncover novel and therapeutically tractable molecular targets that can inspire the development of the next-generation of therapies for these cancers.
 PRMT1 was identified as a top hit in a genome-scale genetic screen for AR regulators. From Tang, Sethunath et al., Cell Reports, 2022
PRMT1 was identified as a top hit in a genome-scale genetic screen for AR regulators. From Tang, Sethunath et al., Cell Reports, 2022
Target discovery in the AR pathway in prostate cancer:
Via genome-scale genetic screening in models of prostate cancer, we recently identified the protein arginine methyltransferase 1, PRMT1, as a novel regulator of androgen receptor signaling and a potential therapeutic target in prostate cancer. This provides preclinical support for the testing of PRMT inhibitors together with AR antagonists in advanced prostate cancer.
We are applying similar approaches to systematically dissect the vulnerabilities of other genitourinary cancers.
Sex Bias in Cancer
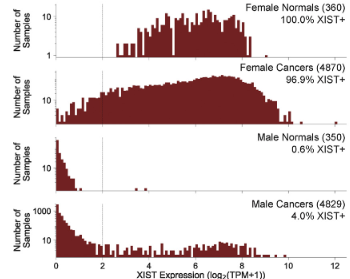 XIST is somatically expressed in a subset of male cancers. From Sadagopan et al., Cell Systems, 2022
XIST is somatically expressed in a subset of male cancers. From Sadagopan et al., Cell Systems, 2022
Both prostate cancer and tRCC share the feature of having a strong driver oncogene on chromosome X. An emerging interest in our laboratory is in using both of these cancers as models to understand how genetic alterations – specifically on the sex chromosomes – may underlie sex differences in cancer incidence or pathogenesis.
Through a pan-cancer genomic analysis, we recently reported that a subset of male cancers can somatically activate the expression of XIST, a normally female-exclusive transcript responsible for dosage compensation of genes on the X chromosome. In ongoing work, we are functionally studying the implications of somatic XIST activation in males and are employing genomic analyses to understand how X-chromosome inactivation can be somatically dysregulated in cancer.
Through a pan-cancer genomic analysis, we recently reported that a subset of male cancers can somatically activate the expression of XIST, a normally female-exclusive transcript responsible for dosage compensation of genes on the X chromosome. In ongoing work, we are functionally studying the implications of somatic XIST activation in males and are employing genomic analyses to understand how X-chromosome inactivation can be somatically dysregulated in cancer.
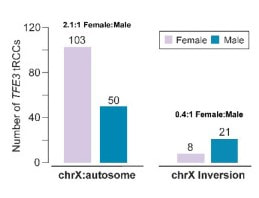 The female predominance of tRCC dominance is largely caused by X:autosomal translocations with a near exact 2:1 female-to-male ratio. From Achom, Sadagopan, Bao et al., bioRxiv, 2023
The female predominance of tRCC dominance is largely caused by X:autosomal translocations with a near exact 2:1 female-to-male ratio. From Achom, Sadagopan, Bao et al., bioRxiv, 2023
By analyzing whole genomes from tRCC, we found that TFE3 rearrangements occur via distinct mechanisms in males and females, with females displaying a 2:1 bias in rearrangements arising via chrX:autosome rearrangements. This ascribes a genetic basis to the female sex bias in tRCC and serves as a model for studying how somatic genetic alterations of the X chromosome may lead to sex-specific constraints on cancer genome evolution.
HOME | RESEARCH | PEOPLE | PUBLICATIONS | SUPPORT | CONTACT
©2019 Viswanathan Lab at Dana-Farber Cancer Institute
©2019 Viswanathan Lab at Dana-Farber Cancer Institute